CASE 2: 17-0036 (4101204-00)
Signalment: 2-month-old male guinea pig (Cavia porcellus).
History: This animal had been received from a commercial supplier several weeks previously and was experimentally unmanipulated. It had been clinically normal, though generally nervous
and skittish until that morning, when it was found recumbent, semi-moribund and in severe
distress, with possible pain when the abdomen was palpated. It expired spontaneously shortly before the planned sacrifice and necropsy.
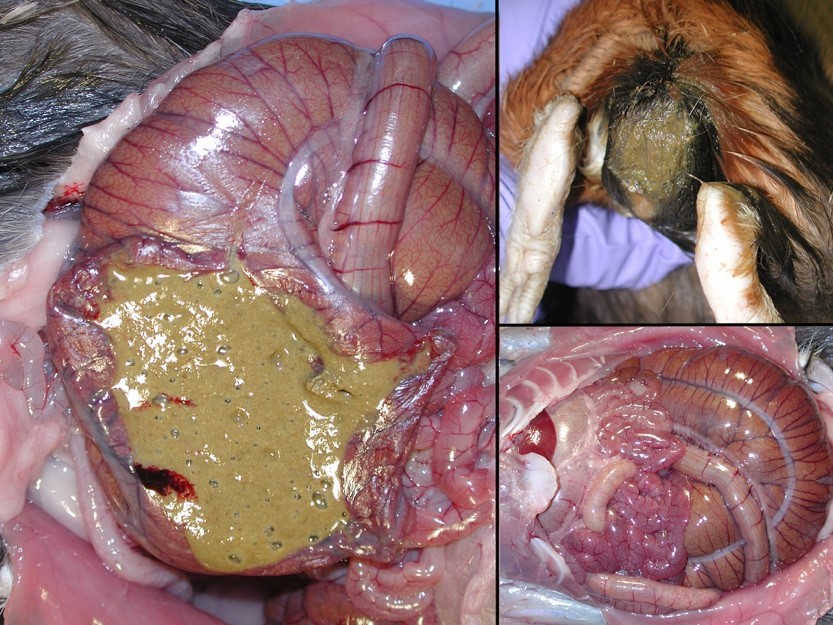
Gross Pathology: The perineum was soiled, and cecum and large intestines were moderately distended with greenish soft to liquid feces.
Laboratory results: Fecal culture for enteric pathogens was negative. Fecal examination for Cryptosporidium and Eimeria were negative. PCR for C. difficile toxin was negative.
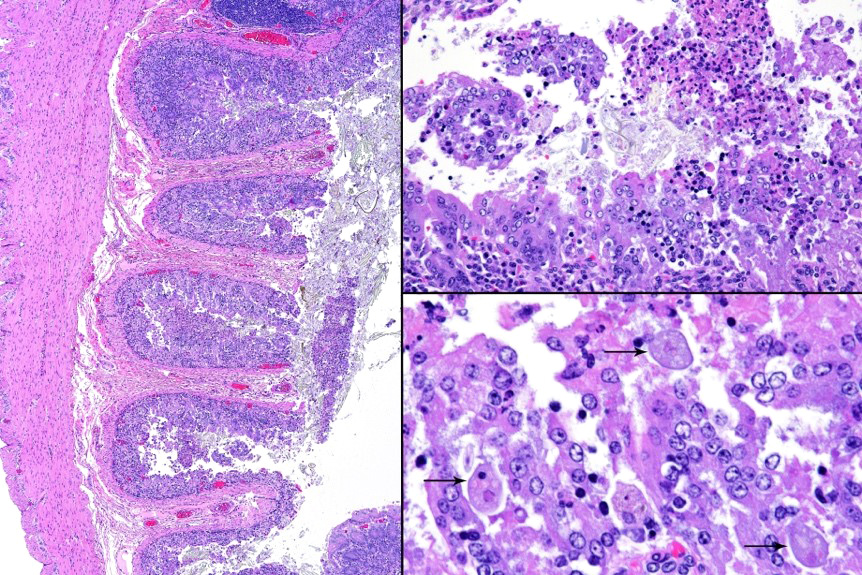
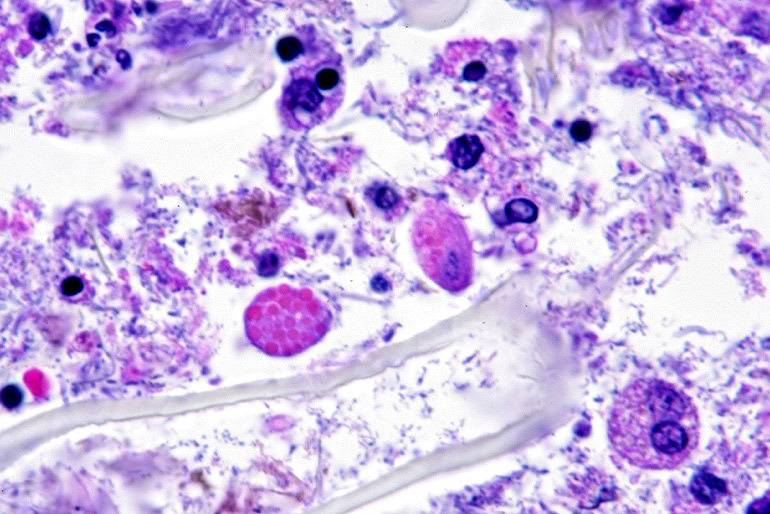
Microscopic description: Full thickness pieces of cecum and colon are examined. There is a patchy to near diffuse (depending on section) acute necrotizing and ulcerative process present, involving primarily superficial mucosal regions, including enterocyte degeneration & exfoliation along with a florid granulocytic infiltrate. Inflammation extends into deeper mucosal regions in many areas as well. Admixed with superficial epithelial and luminal necrotic debris, but also seen extending into deeper mucosal regions are numerous protozoan organisms. These are consistent morphologically with both cyst and trophozoites forms of Entamoeba. Erythrophagocytosis is frequently present within trophozoites structures (abundance of red cell ingestion not evident in all sections).
Contributor's morphologic diagnosis:
Typhlocolitis, necrotizing and ulcerative, acute, patchy to diffuse, marked, with numerous surfaces and more deeply invasive protozoal organisms consistent with Entamoeba sp. (Observed both in tissue section and on wet mount fecal smears).
Contributor's comment: Entamoeba infection and disease (amoebiasis) in mammalian species primarily affects humans and other primates and is generally associated with Entamoeba histolytica. Other mammals such as cats and dogs can occasionally be spontaneously infected with this organism although clinical disease is rare. Amoebiasis in reptiles, typically associated with Entamoeba invadens is well recognized, being commonly reported in snakes and lizards. In search of animal models of disease, rats, mice, guinea pigs, and rabbits have been experimentally infected, typically by injecting large numbers of trophozoites directly into the lower bowel per annuum, or surgically introducing them into the cecum.5 However, rodents appear to be naturally resistant to infection, the cellular and molecular basis of such immunity not being completely understood.4
Innate Entamoeba species have been identified in a variety of rodents and are generally considered to be commensal inhabitants of the cecum; Entamoeba muris is well described in rats, mice and golden hamsters.2
Entamoeba cobayae was described in guinea pigs by Walker in 1908, although this is now considered synonymous with the more commonly designated E. caviae.3 Indeed E. caviae is known to be a common inhabitant of the laboratory guinea pig and is widely considered to be non-pathogenic.6
In earlier days, Entamoeba were identified and classified on the microscopic basis of nuclear structure of trophozoites and cysts found in stool preparations.3 The basis for such identifications depended greatly on the skill and expertise of the microscopist and previously, no means were available to confirm results other than reexamination of the sample by a more experienced microscopist. Furthermore, morphologically identical species or genetic variants could not be distinguished solely on this basis.14 Apart from size, number and appearance of the nuclei, chromatoid bar appearance and several other features, there are few criteria to differentiate between organisms and the use of minor morphological differences to separate species may turn out to be not reliable.7 A case in point involves Entamoeba dispar; a separate but non-invasive and non-pathogenic Entamoeba species in humans which is microscopically indistinguishable from E. histolytica and does not require treatment.7,13
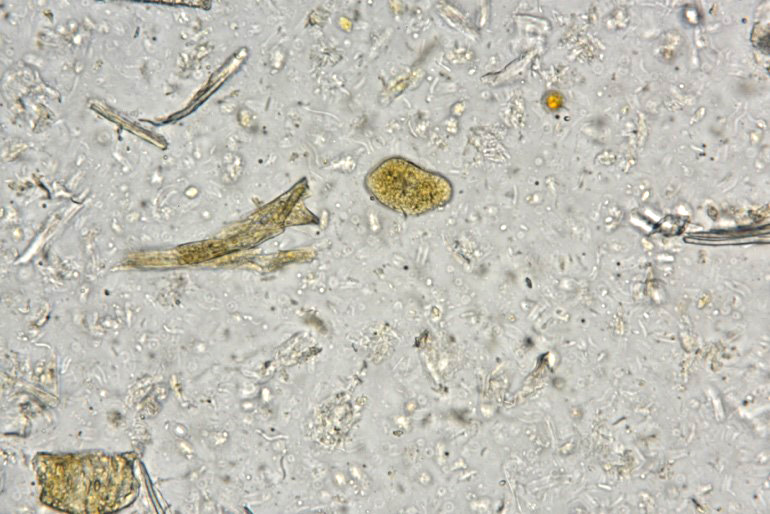
Examination of fecal wet mounts in this case revealed abundant trophozoites structures compatible with Entamoeba generally measuring between ~20 ? 26 µm in diameter, with nuclei ~5.0 ? 6.2 µm possessing a ring of coarse peripheral granules inside the nuclear membrane. Cysts were present, though rare, measuring ~20 µm in diameter. Trichrome staining is pending to try and identify the number of nuclei present, as this can be an important consideration in distinguishing between pathogenic and non-pathogenic types of the organism.2
Alternative means of identification beyond morphology include isoenzymatic methods, immunological analysis (both antibody and antigen detection) and molecular diagnostics (both conventional and Real-time PCR).9 Certainly the majority of molecular diagnostic tests available focus on distinguishing E. histolytica, E. dispar and other non-pathogenic species known in humans.
Initial PCR fecal testing on this animal was negative for E. histolytica, E. dispar and E. moshkovskii), ruling these organisms out as etiologic considerations (though they were not thought to be likely agents based on historical knowledge of their host range). Additional frozen samples from the case were submitted to a laboratory for DNA sequencing. Results indicated that the organism appears to be an uncharacterized species of Entamoeba that is most closely related to Entamoeba coli, with ~97% homology. Unfortunately, Entamoeba caviae does not have sequence data available in GenBank and therefore could not be completely excluded based on known genomic standards.
The possibility that the primary necrotizing enteric disease present in this case was of another etiology and that the amoebic organisms present had proliferated in response to this was considered, but thought less likely due to 1) the presence of deep invasion of organisms into the mucosa, 2) the negative culture and PCR results for other known causes of typhlocolitis in Guinea Pigs and 3) the lack of morphological evidence of other etiologic agents such C. piliforme via special stains. Another indirect consideration suggesting a primary Entamoeba pathogenesis in this case is the high degree of erythrophagocytosis noted in section. Phagocytic activity by pathogenic Entamoeba trophozoites (including E. histolytica) has been accepted as a qualitative pathogenicity factor and one associated with virulence mechanisms responsible for invasive capacity.10
In summary, despite the lack of substantial similar reports in the literature, this case of necrotizing typhlocolitis was considered likely of a primary Entamoeba etiology, although other infectious cofactors could not be completely excluded. The exact species of Entamoeba involved was undetermined, as was the reason for the susceptibility of this animal to infection.
Contributing Institution:
Division of Laboratory Animal Resources
University of Pittsburgh
www.dlar.pitt.edu
JPC diagnosis:
Colon: Colitis, necrotizing, multifocal, moderate, with moderate numbers of extracellular amoebic trophozoites, guinea pig, rodent.
JPC comment:
The contributor highlights important background information for this entity, as well as obstacles sometimes encountered in veterinary medicine. As computational pathology matures as a field, we expect diagnostic capabilities to increase dramatically. Perhaps sequences for E. caviae will be available in the near future.
In humans and non-human primates (NHP), the primary sites of dissemination are the liver (via the portal circulation), and less commonly, the lung and brain. Fatal amebiasis with abscess formation has been reported in multiple primate species.8
The pathogenesis of tissue damage caused by amebae species are:
1. Adhesion to mucus by lectins
2. Enzymatic breakdown of protective mucus
3. Lectin-mediated adherence to host epithelium
E. histolytica releases cysteine proteases that cause damage to mucosal epithelium and attract inflammatory cells, both of which lead to characteristic ulcerative colitis with flask-shaped ulcers. Microscopically, amebae are surrounded by a clear halo with extensive pseudopodia and possess a nucleus with a dark karyosome and peripheral chromatin clumps. The cytoplasm appears foamy and they frequently phagocytize erythrocytes, which makes them difficult to distinguish from activated macrophages. Their cytoplasm often contains glycogen, making amebae PAS positive.13
Amoebae continue to be important zoonotic pathogens of concern, and have been found in wild and captive prosimians, apes (Balamuthia mandrillaris, E. histolytica, E. dispar, Dientamoeba fragilis, and other members of Entamoeba spp, Dientamoeba spp, and Iodoamoeba sp.), reptiles (E. invadens), amphibians (E. ranarum), and domestic pigs (D. fragilis). Interestingly, given its aquatic environment, captive tapir in South America have developed necrosuppurative meningo-encephalitis from infection by Naegleria fowleri.12 This amoeba is often known as the ?brain-eating amoeba? and infects humans by extension from nasal passages and through the cribriform plate, causing primary amebic meningoencephalitis (PAM). Death is acute, and little inflammation develops.15
Recent molecular investigations have elucidated the variety of Entamoeba species that infect humans and nonhuman primates. Leveraging current techniques using monoclonal antibodies, DNA hybridization, SSU-rDNA restriction fragment length polymorphism, and DNA sequencing, a distinct species infecting nonhuman primates was isolated from previously presumed E. histolytica cases and named Entamoeba nuttalli. In total, there are six named Entamoeba species that infect nonhuman primates (E. coli, E. polecki, E. histolytica, E. nuttalli, E. dispar, and E. hartmanni), as well as four that are not yet named, but identified by unique gene sequences (Entamoeba RL3, RL6, RL7, and CL8).1,11
References:
1. Elsheikha HM, Regan CS, Clark CG. Novel Entamoeba findings in nonhuman primates. Trends in Parasitology. 2018;34(4):283-294.
2. Fox J, Barthold S, Davisson M, Newcomer C, Quimby F, Smith A. The Mouse in Biomedical Research 2nd Edition: Volume II Diseases, New York: Academic Press; 2007: 527-528.
3. Hooshyar H, Rostamkhani P, Rezaeian M. An Annotated Checklist of the Human and Animal Entamoeba (Amoebida: Endamoebidae) Species ? A Review Article. Iranian J Parasitol 2015; 10:146-156.
4. Jarillo-Luna R, Campos-Rodriguez R, Tsutsumi V. Entameoeba histolytica: Immunohistochemical study of hepatic amoebiasis in mouse. Neutrophils and nitric oxide as possible factors of resistance. Experimental Parasitology; 2002 101: 40-56
5. Jervis H, Takeuchi A. Amebic dysentery. Animal model: experimental Entamoeba histolytica infection in the germfree guinea pig. Am J Path; 1979 94(1): 197-200.
6. Nie D. Morphology and taxonomy of the intestinal protozoa of the Guinea-pig, Cavia porcella. Journal of Morphology. 1950; 86: 331-493.
7. Nozaki T, Bhattacharya A (eds), Amebiasis: Biology and Pathogenesis of Entamoeba, Japan: Springer; 2015:21-22.
8. Strait K, Else JG, Eberhard ML. Parasitic diseases of nonhuman primates. In: Abee CR, Mansfield K, Tardif S, Morris T, ed. Nonhuman Primates in Biomedical Research: Diseases. Vol 2. 2nd ed. San Diego, CA: Elsevier; 2012:206-209, 221-222, 599-602.
9. Subhas C, Mandai J, Ponnambath D. Laboratory methods of identification of Entamoeeba histolytica and its differentiation from look-alike Entamoeba spp. Trop Parasitol 2014; 4(2): 90-95.
10. Talamas-Lar D, Chavez-Munguia B, Gonzalez-Robles A, Talamas-Rohana P, Salazar-Villatoro L, Duran-Diaz A, Paloma-Martinez A. Erythrophagocytosis in Entamoeba histolytica and Entamoeba dispar: A Comparative Study. BioMed Research International. June 2014 DOI: 10.1155/2014/626259
11. Tanaka M, Makiuchi T, Komiyama T, et al. Whole genome sequencing of Entamoeba nuttalli reveals mammalian host-related molecular signatures and a novel octapeptide-repeat surface protein. Neglected Tropical Diseases. 2019;13(12):e0007923.
12. Terio KA, McAloose D, St. Leger J. Pathology of Wildlife and Zoo Animals. San Diego, CA: Elsevier; 2018.
13. Uzal FA, Plattner BL, Hostetter JM. Alimentary system. In: Maxie, MG, ed. Jubb, Kennedy, and Palmer's Pathology of Domestic Animals. Vol 2. 6th ed. St. Louis, MO: Elsevier; 2016:242.
14. Verweij JJ, Laeijendecker D, Brienen E, Lieshout L, Polderman A. Detection and Identification of Entamoeba Species in Stool Samples by a Reverse Line Hybridization Assay. Journal of Clinical Microbiology. 2003; 41(11): 5041 ? 5045.
15. Visvesvara GS. Free-living amebae as opportunistic agents of human disease. J Neuroparasitol. 2010;1.