Joint Pathology Center
Veterinary Pathology Services
Wednesday Slide Conference
2018-2019
Conference 13
2 January, 2018
CASE I: A-475-4 (JPC 4100984).
Signalment: 4-8 week old female C57BL/6NCrl mouse (Mus musculus)
History: Experimentally infected with mouse adapted Ebola virus (ma-EBOV) by intraperitoneal route
Gross Pathology: Gross lesions included a pale and friable liver, pale kidneys, enlarged spleen and hemorrhagic/congested gastrointestinal tract
Laboratory results: N/A
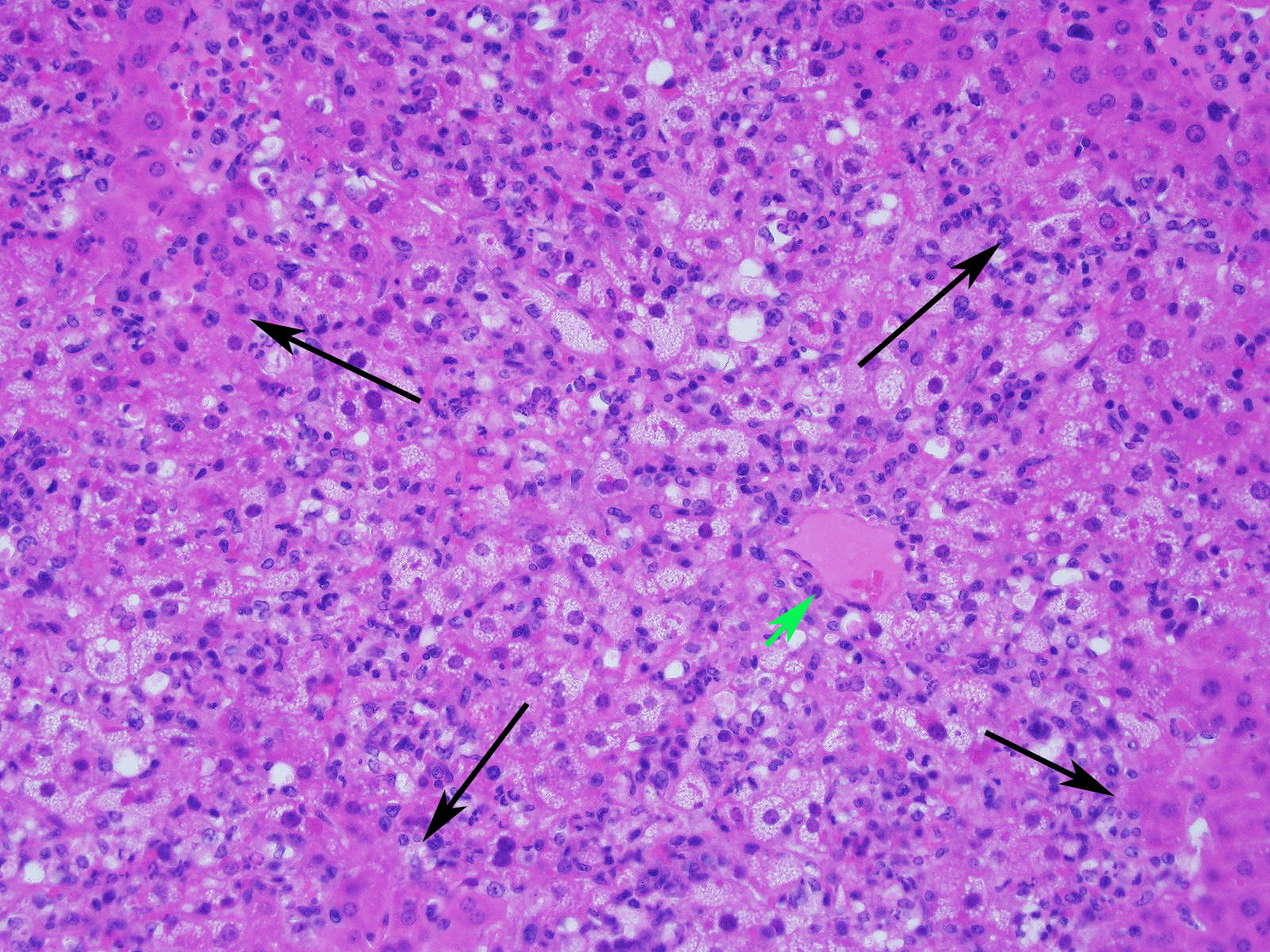
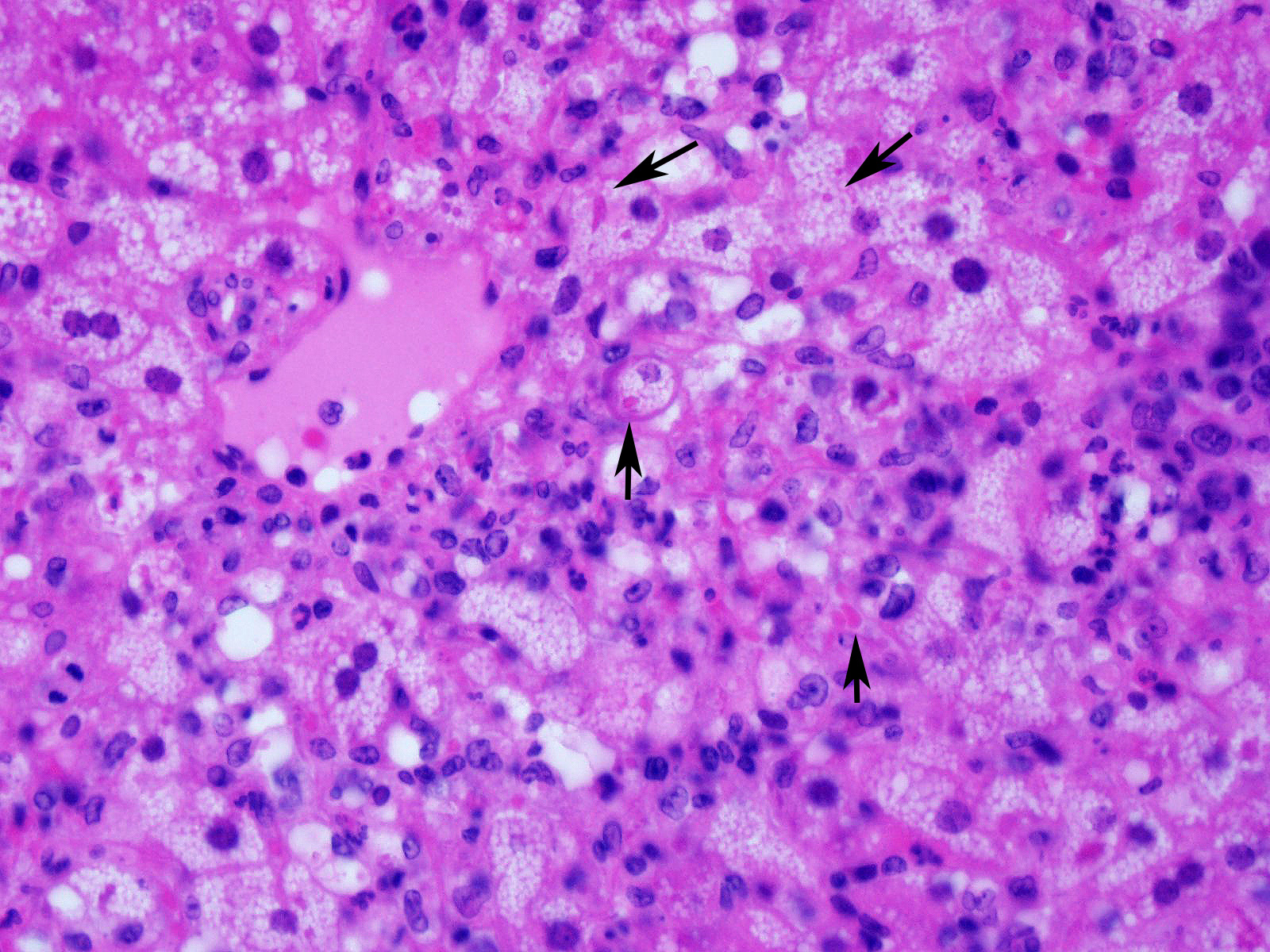
Microscopic Description:
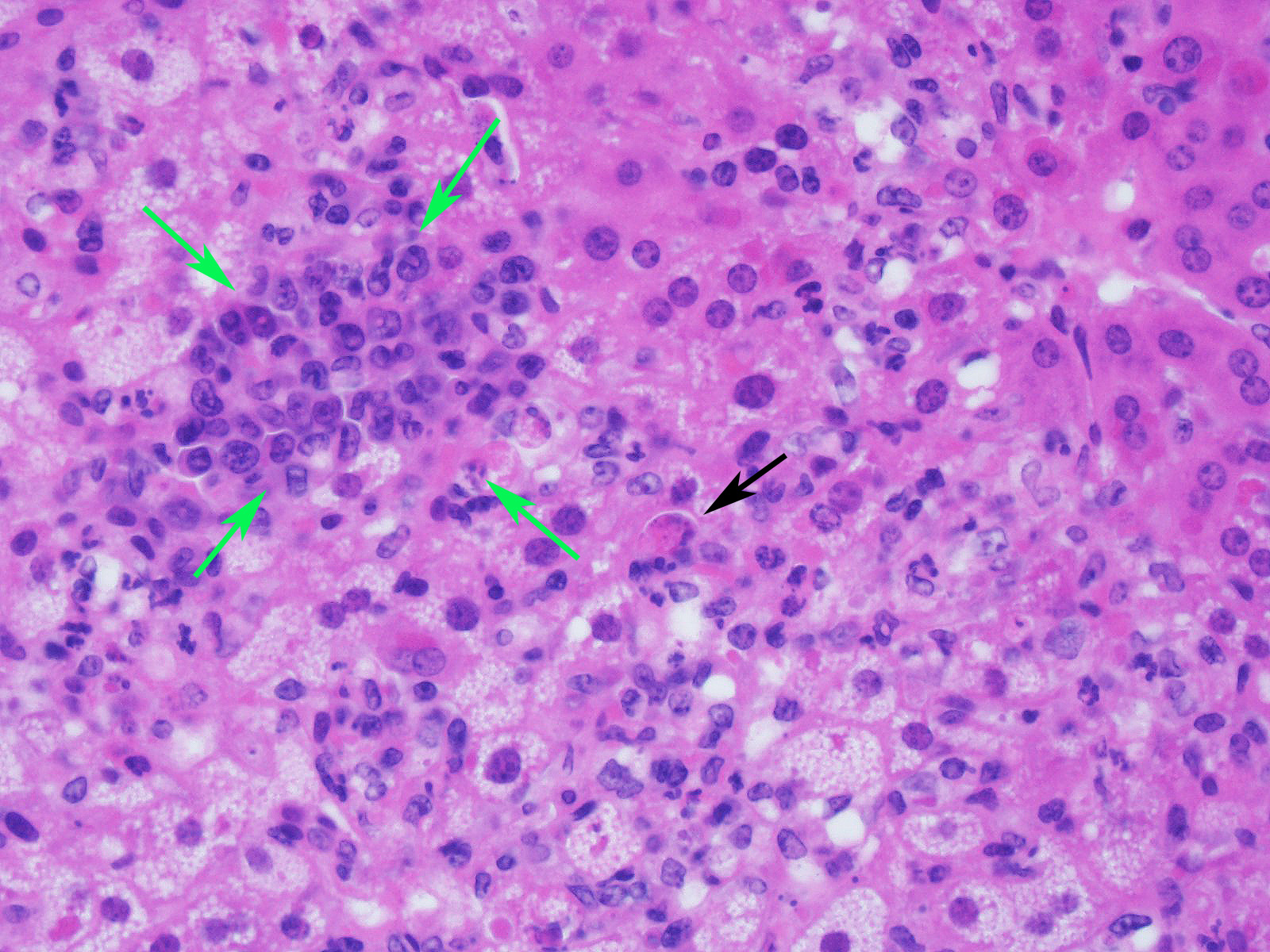
Slide contains a single section of liver. There is severe multifocal to coalescing predominantly periportal and midzonal hepatocellular degeneration and necrosis with moderate to severe mixed inflammation. In approximately 30% of lobules there is massive panlobular degeneration and inflammation. There is abundant periportal to midzonal microvesicular lipid type cytoplasmic vacuolation of hepatocytes, with frequent hyaline eosinophilic cytoplasmic inclusions (resembling Councilman bodies) and cell swelling. There are several foci of hepatocyte dissociation and loss with acute hemorrhage. Apoptotic and necrotic hepatocytes are common. Small to large eosinophilic intracytoplasmic (viral) inclusions are present within viable and degenerate hepatocytes, Kupffer cells and rarely endothelial cells. Inflammatory cell infiltrate includes large numbers of macrophages and viable and degenerate neutrophils. Within central veins there are frequent vacuolated monocytes, occasionally with phagocytosed apoptotic debris or eosinophilic intracytoplasmic inclusions. There are scattered linear to branching fine olive green bile plugs within canaliculi between hepatocytes.
Contributor’s Morphologic Diagnoses:
Liver, hepatitis, necrotizing and histiocytic, multifocal to coalescing, acute, severe with intracytoplasmic inclusion bodies
Contributor’s Comment: Liver lesions are severe and consistent with experimental mouse-adapted Ebola virus infection.2,3 Ebola virus, a filovirus, is an important high consequence pathogen causing significant human disease, most recently a large outbreak in west Africa from 2014 to 2016, with a smaller outbreak in 2017. Ebola targets hepatocytes and Kupffer cells of the liver, resulting in widespread hepatocellular degeneration and necrosis. Liver lesions in human and non-human primate infections are conspicuous for the minimal to absent leukocytic infiltrate, in contradistinction to the lesion presented here.1,7
The extensive inflammation and absence of a distinct zonal (lobular/acinar) pattern point toward an infectious/inflammatory rather than toxic etiology. Not knowing the strain of mouse, immune status, experimental manipulation or other tissue lesions, reasonable differential etiologies for severe necrotizing hepatitis in a (usually immunodeficient) mouse would include mouse hepatitis virus (MHV, type species of Betacoronavirus), mouse adenovirus (MAdV-1 or -2, Mastadenovirus), mouse cytomegalovirus (MCMV, murid betaherpesvirus-1, type species of Muromegalovirus), lymphocytic choriomeningitis virus (LCMV, type species of Mammarenavirus), or ectromelia virus (mousepox, Orthopoxvirus).1 Mouse adenovirus and MCMV have intranuclear rather than intracytoplasmic inclusions, and mouse hepatitis virus lacks inclusions but forms characteristic syncytia. Ectromelia does form cytoplasmic inclusions in hepatocytes, but these tend to be difficult to visualize without prolonged hematoxylin staining. Viral cytoplasmic inclusions in hepatocytes must also be differentiated from phagocytosed neighboring apoptotic hepatocytes (Councilman bodies). Acute salmonellosis and Tyzzer’s disease would be additional differential etiologies, although the lesions tend to be more nodular, with intracellular bacterial stacks in the latter.1
Contributing Institution:
Pathology Department
NIH/NIAID
Integrated Research Facility
https://www.niaid.nih.gov/about/integrated-research-facility
JPC Diagnosis: Liver: Hepatitis, necrotizing and neutrophilic, random, diffuse,
severe, with numerous intracytoplasmic viral inclusions.
JPC Comment: The first Ebola virus outbreak occurred in the Democratic Republic of the Congo (DRC) in 1976, resulting in 280 cases as a result of close contact and use of contaminated needles. A large outbreak in Western Africa from 2013-2015 claimed over 11,000 lives in multiple countries, with the majority of deaths in Sierra Leone, Liberia and Guinea with cases exported to other countries in Europe and North America for the first time.4 At the time of this writing, another outbreak occurs in the DRC, with 130 confirmed cases. Outbreak control has been the key to the decreased in lives claimed by this disease over the last forty years, effective treatment and vaccination for this hemorrhagic fever of humans is still woefully inadequate.
The genus Ebolavirus is divided into five distinct species: Zaire ebolavirus (EBOV), Sudan ebolavirus, Tai Forest ebolavirus, Bundibugyo ebolavirus, and Reston ebolavirus (a species of ebolavirus that is fatal in macaques but not in humans.) The ebolaviruses can infect a number of animal species including non-human primates (chimpanzees, monkeys, macaques, orangutans, baboons), fruit bats, dogs, rodents, porcupines, and a range of laboratory rodents.4 During human epizootics, bats have served as reservoirs in the source for infection. Bats are subclinical carriers, and do not show clinical signs of infection. Studies on arthropods have shown that they are not vectors of EBOV.4
Shortly after the initial outbreak in the DRC, nonhuman primate and guinea pig models were developed for infection with Zaire ebolavirus. The hemorrhagic fever seen in nonhuman primates closely resemble the human disease.2 However the complications of working with nonhuman primate models, as well as their expense, demonstrated the need for the development of murine models. In addition, the possibility of using genetically engineered murine models to study specific cytokine interactions in a very complex viral pathogenesis further reinforced a call for development of an appropriate murine model. Ultimately, an appropriate murine model was eventually developed by passaging Zaire ebolavirus from the original outbreak through progressively older suckling mice until it could be transmitted to adult, immunocompetent BALB/c mice.2
Mouse models such as the BALB/C share many similarities with guinea pig and NHPS models with cells of the mononuclear phagocyte system being target by the virus early in the disease. Viral replication may be seen in macrophages in lymph nodes and the spleen within 2 days of infection. On day three, when mice first show signs of clinical illness, virus replication is widespread, with infection of hepatocytes, adrenocortical cells, fibroblasts, adipocytes, and cells of the monocyte-macrophage system in all organs. Histologic changes on day five include marked lymphocytolysis in multiple organs. Endothelial cell infection, even in late stages of disease, is uncommon. One notable difference in the mouse model as compared to humans and the NHP model is the noticeable paucity of fibrin thrombi within the spleen of infected mice (a very prominent finding in other model species as well as humans.)2
Since the development of the first murine model, flaws in the model have become apparent, as mouse models do not display other consequences of disease noted in humans, such as disseminated intravascular coagulation or death from shock, as seen in non-human primates. (An excellent 2015 article by Martines et al.5 from the Centers for Disease Control and Prevention reviews the disease caused by ebolaviruses and the related Marburg virus in humans). Since that time, research in a wide variety of inbred mouse strains have identified resistant and susceptible strains and potentially responsible resistance genes which may reflect the apparent genetic susceptibility displayed in humans when a large population is infected by the virus.6
The moderator noted the difference between Ebola infections in mice and in primates, with the almost total lack of inflammatory cells within primate specimens. Ebola is not a virus that can be simply injected in mouse models – it is serially passaged, and mouse-adapted strains need to be injected IP for infection. He also discussed the peculiar abnormalities associated with C57BL/6N strains, including retinal dysplasia, and if they are 6Nhsd, a mutation which causes an impaired immune response.
The moderator cautioned about overinterpreting the lipid change in this particular animal – it may be quite variable based on time of day, elapsed time since last meal, and in sick mice, inability to feed.
References:
- Barthold, S.W., S.M. Griffey, and D.H. Percy, Mouse, in Pathology of Laboratory Rodents and Rabbits, S.W. Barthold, S.M. Griffey, and D.H. Percy, Editors. 2016, Wiley Blackwell: Ames, IA. p. 1-118.
- Bray M, Davis K, Geisbert T, Schmaljohn C, Huggins J. A mouse model for evaluation of prophylaxis and therapy of Ebola hemorrhagic fever. J Infect Dis 1998; 178(3): 651-61.
- Gibb TR, Bray M, Geisbert TW, Steele KE, Kell WM, Davis KJ, Jaax NK. Pathogenesis of experimental Ebola Zaire virus infection in BALB/c mice. J Comp Pathol 2001. 125(4):233-42.
- Gumusova S, Sunbul M, Lebebicioglu. Ebola virus disase and the veterinary perspective. Ann Clin Microbiol and Antimicrobials 2015; 14:30-35.
- Martines RB, Ng DL, Greer PW, Rollin PE, Zaki SR. Tissue and cellular tropism, pathology and pathogenesis of Ebola and Marburg viruses. J Pathol 2015. 235(2):153-74.
- Rasmussen AL, Okumura A, Ferris MT, Green R, Feldmann F, Kelly SM, Scott DP, Safronetz D, Haddock E, LaCase R, Thomas MJ, Sova P, Carter VS Weiss JM, Miller DR, Shaw GD, Korth MJ, Heise MY Baris RS, deVilena FPD, Feldmann H, Katze MG. Host genetic diversity enables Ebola hemorrhagic fever pathogenesis and resistance. Science 2014; 346(6212):987-991.
- Ryabchikova, E.I., L.V. Kolesnikova, and S.V. Luchko, An analysis of features of pathogenesis in two animal models of Ebola virus infection. J Infect Dis 1999; 179 Suppl 1: S199-202.